Mylotools overview
Mylotools is a set of utilities for working with myloasm assemblies. Source code for mylotools can be found here.
The main use of mylotools is to visualize assembled contigs for quality control. There are also functions for processing myloasm's contigs.
Mylotools currently has the following useful functionality:
usage: mylotools [-h] [--version] {report,sanitize-headers,plot,strain-viz,extract-contigs} ...
positional arguments:
{report,sanitize-headers,plot,strain-viz,extract-contigs}
report Generate comprehensive report with plots for all long contigs
sanitize-headers Sanitize FASTA headers by replacing underscores with spaces (creates backup)
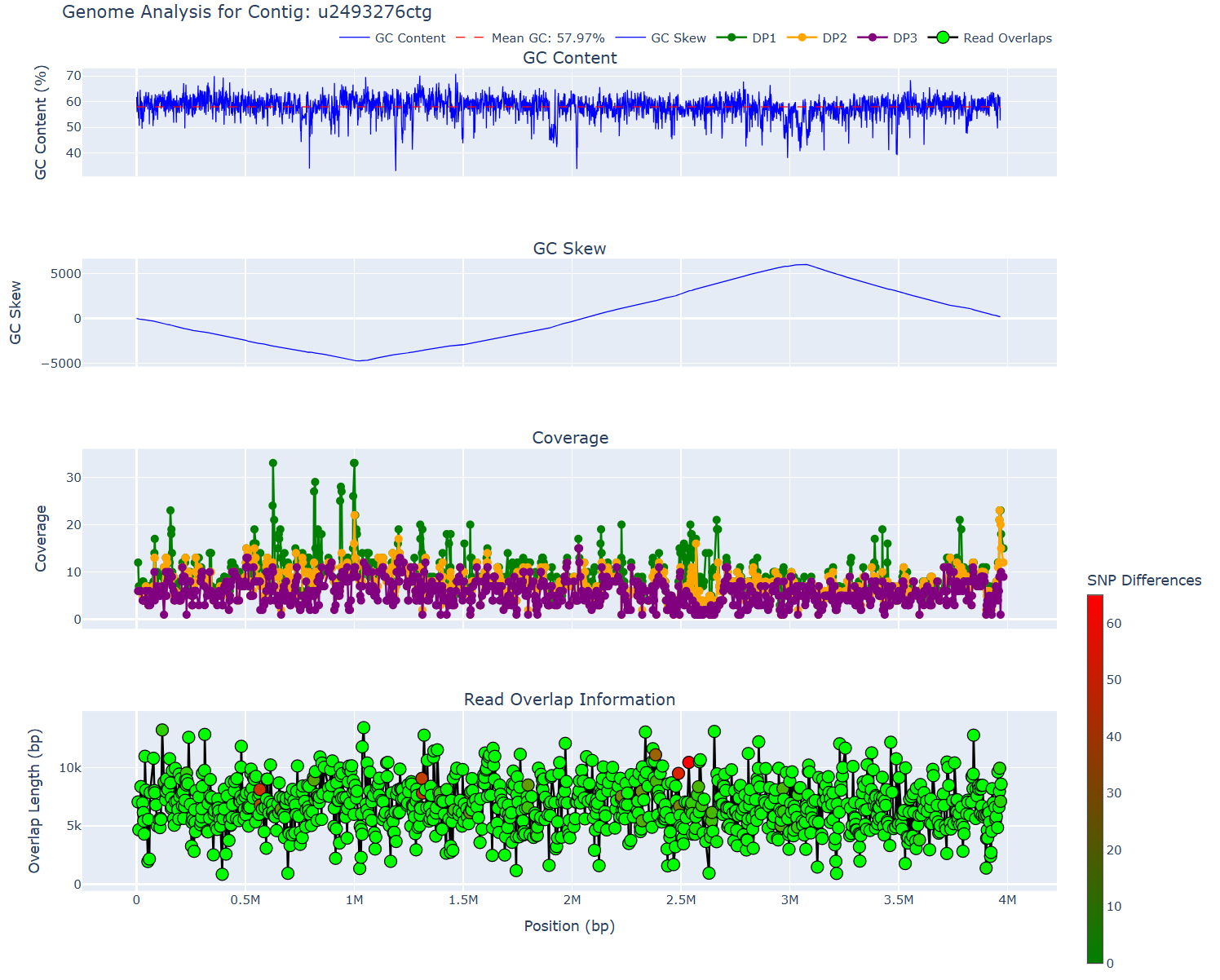
plot Generate a plot of various statistics for one of myloasm's output contigs (`report` does `plot` for all contigs)
strain-viz Visualize overlaps between and within two or more similar contigs
extract-contigs Extract contigs > X bp into a folder as its own fasta
Installing mylotools
Conda install
mamba install -c bioconda mylotools
Clone + pip
git clone https://github.com/bluenote-1577/mylotools
cd mylotools
## assuming you have python3 installed
pip install .
Using mylotools
It's easiest to use mylotools by cd-ing into the assembly output.
myloasm reads.fq .... -o myloasm_results
cd myloasm_results
Generating an HTML report
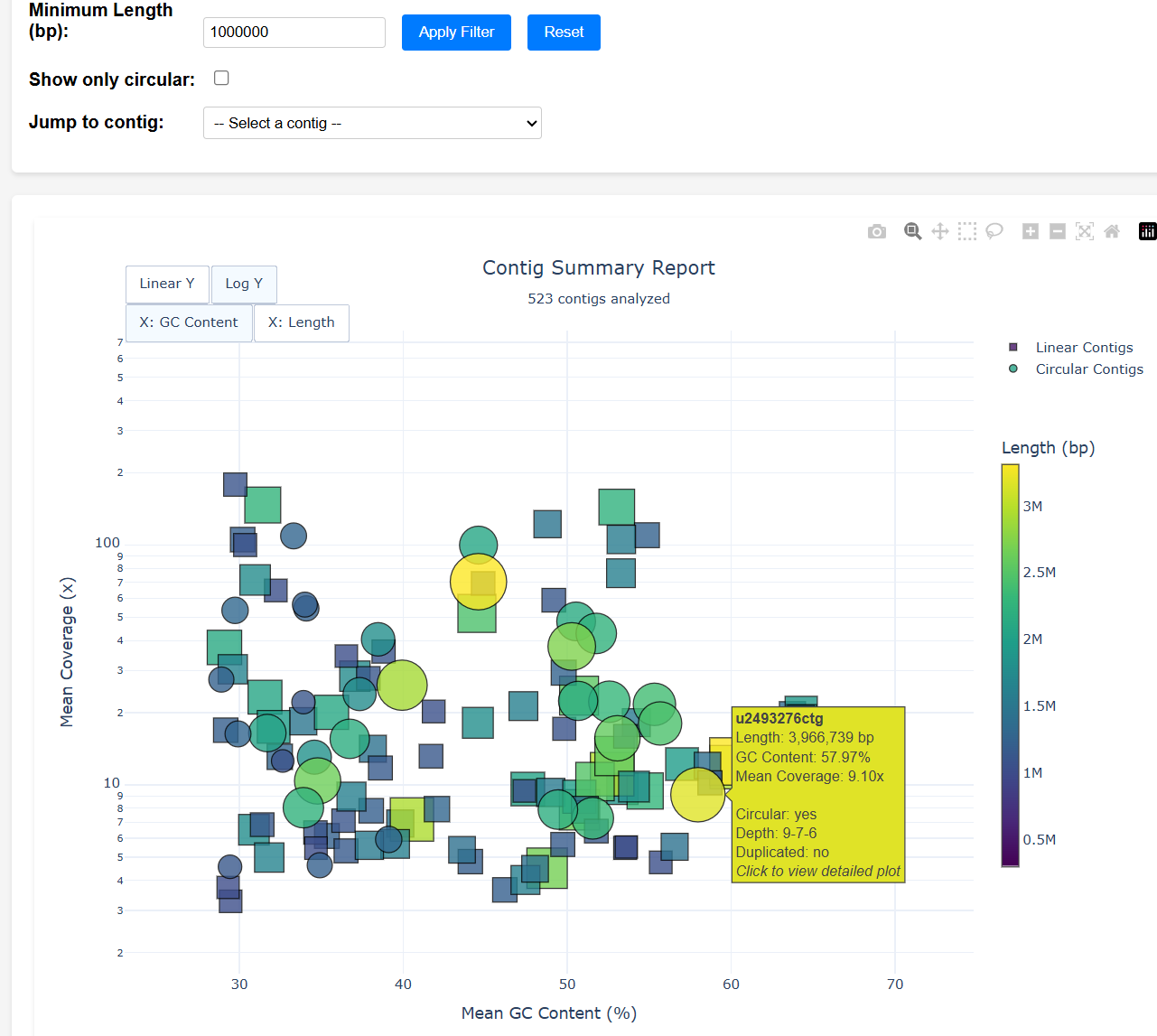
mylotools report -o mylo-report
ls mylo-report/contig_summary_report.html
This extracts all contigs of length > 300 kbp OR designated circular (by default) and then runs mylotools plot to generate QC plots. contig_summary_report.html is an interactive plot -- clicking a contig redirects to its QC plot.